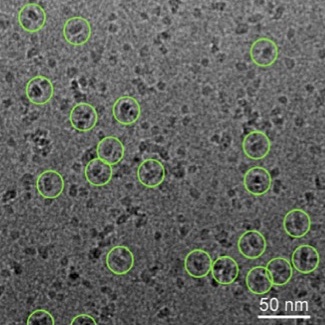
Cryo-electron microscopy images of membrane proteins (circled). Image courtesy of our collaborator Cornelius Gati.
Protein Shapes
Proteins are fundamental to life with functions ranging from mechanical cellular support to immune protection. The specific role of a given protein is mainly determined by its 3D shape, which is why it is so crucial to understand proteins' 3D conformations.
Our research implements and uses tools from geometric learning to:
- reconstruct the shapes of proteins from biological images such as images generated by cryo-electron microscopy,
- analyzes the protein shapes in relation with their biological functions
Our current research focuses on membrane proteins, these molecules that are the target of more than 50% of prescription drugs, including the treatment of neurological disorders and cancers, yet still difficult to image.
Want to learn more?
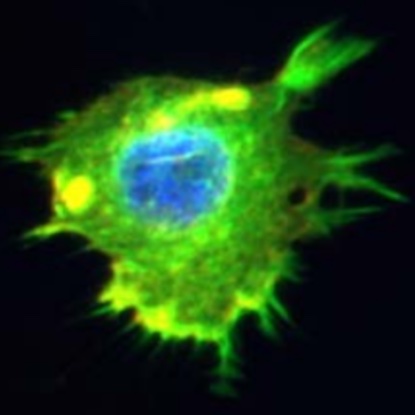
Irregular boundary of a cancer cell in the mouse. Image courtesy of our collaborator Ashok Prasad.
Cell Shapes
Cells adopt a variety of shapes determined by the biophysical forces and processes at the very center of life. The emergence of large-scale cell imaging gives us a fantastic opportunity to study these shapes with quantitative measures that link cellular morphology with cellular functionality and cellular health.
Our research implements and uses tools from geometric learning to:
- perform quantitative analyses of cell shapes under different physiological conditions,
- link the geometry of the cell to its biophysical processes.
Our current research studies how shape differences between cancer cells treated with different drugs can shine light on the underlying biophysical processes.
Want to learn more?
- Check out this notebook analyzing cancer cell lines with geometric methods!
- Look at the research from our collaborator Khanh Dao Duc!
Relevant Publications
2022
- Gabbert, A., Mondo, J., Campanale, J., Mitchell, N., Myers, A., Streichan, S., Miolane, N., Montell, D. "Septins Regulate Border Cell Shape and Surface Geometry Downstream of Rho." || Journal Developmental Cell (2022 - in review).
- Donnat, C., Levy, A., Poitevin, F., Miolane, N. "Deep Generative Modeling for Volume Reconstruction in Cryo-Electron Microscopy." || Journal of Structural Biology (2022 - in review). [Paper] [Code].
- Myers, A., Miolane, N. "Regression-Based Elastic Metric Learning on Shape Spaces of Elastic Curves." || NeurIPS Workshop on Learning Meaningful Representations of Life. (2022). [Paper] [Code (coming soon)].
- Nashed, Y., Peck, A., Martel, J., Levy, A., Koo, B., Wetzstein, G., Miolane, N., Ratner, D., Poitevin, F. "Heterogeneous Reconstructions of Deformable Models in Cryo-Electron Microscopy." || NeurIPS Workshop of Machine Learning for Structural Biology (2022). [Paper] [Code (coming soon)].
- Levy, A., Poitevin, F., Martel, J., Nashed, Y., Peck, A., Miolane, N., Ratner, D., Dunne, M., Wetzstein, G. "CryoAI: Amortized Inference of Poses for Ab Initio Reconstruction of 3D Molecular Volumes from Real Cryo-EM Images." Best Poster Award at the 4th International Symposium on Cryo-3D Image Analysis. || ECCV European Conference on Computer Vision (2022). [Paper] [Code].
- Utpala, S., Miolane, N. "Biological Shape Analysis with Geometric Statistics and Learning." || Oberwolfach Snapshots of Modern Mathematics (2022).
- Legendre, N, Dao Duc, K., Miolane, N. "Defining an Action of SO(d)-Rotations on Images Generated by Projections of d-Dimensional Objects: Applications to Pose Inference with Geometric VAEs." || GRETSI Conference (2022). [Paper].
Previous
-
Miolane, N., Poitevin, F., Li, Y.-T., Holmes, S. "Estimation of Orientation and Camera Parameters from Cryo-Electron Microscopy Images with Variational Autoencoders and Generative Adversarial Networks." || CVPR Workshop on Computer Vision for Microscopy Imaging (2020). [Paper].
-
Miolane, N., Poitevin, F., Holmes, S. "Exploring Cryo-EM Latent Space with Variational Autoencoders." || Stanford Bio-X Workshop on Cryo-Electron Microscopy. (2019).
-
Poitevin, F., Li, Y.T., Miolane, N., Gati, C., Levitt, M. "Convenience Tools to Explore Variability in Cryo-EM Data." || Stanford Bio-X Workshop on Cryo-Electron Microscopy (2019).